DLRG schedule for autumn semester 2025
(GoogleMeet link: https://meet.google.com/qxw-yxyi-pjf )
1) Sunday (Oct. 12, 2025) presented by Abdallah, “Generating dermatopathology reports from gigapixel whole slide images with HistoGPT”. url: https://doi.org/10.1038/s41467-025-60014-x
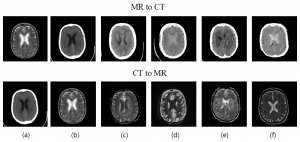
2) Sunday (Oct. 26, 2025) presented by Muna, “A diffusion model for universal medical image enhancement”. url: https://doi.org/10.1038/s43856-025-00998-1
3) Sunday (Nov. 9, 2025) presented by Jana, Either, Dana & Leen, “Generative Multimodal Pretraining with Discrete Diffusion Timestep Tokens”. url: https://arxiv.org/pdf/2504.14666
4) Sunday (Nov. 23, 2025) presented by Rahmeh, “Deep learning‑based cardiac computed tomography angiography left atrial segmentation and quantification in atrial fibrillation patients: a multi‑model comparative study”. url: https://doi.org/10.1186/s12938-025-01442-0
5) Sunday (Dec 7, 2025) presented by Adnan, Basil, Dunia & Ihmaidan “ NoProp: Training Neural Networks without Full Back-propagation or Full Forward-propagation”. url: https://arxiv.org/pdf/2503.24322
6) Sunday (Dec 21, 2025) presented by Alaa, “DeMamba: AI-Generated Video Detection on Million-Scale GenVideo Benchmark”, url: https://arxiv.org/abs/2405.19707
7) Sunday (Jan. 4, 2026) presented by , “”. url:
(GoogleMeet link: https://meet.google.com/qxw-yxyi-pjf )
2) Sunday (Mar. 23, 2025) presented by Besan, Liyan, Leen and Zaina, “Storynizor: Consistent Story Generation via Inter-Frame Synchronized and Shuffled ID Injection”. url: https://arxiv.org/pdf/2409.19624
3) Sunday (Apr. 6, 2025) presented by Fatima, “HiFi-Syn: Hierarchical granularity discrimination for high-fidelity synthesis of MR images with structure preservation”. url: https://doi.org/10.1016/j.media.2024.103390
4) Sunday (Apr. 20, 2025) presented by Abdullah, “Ensemble transformer-based multiple instance learning to predict pathological subtypes and tumor mutational burden from histopathological whole slide images of endometrial and colorectal cancer”. url: https://doi.org/10.1016/j.media.2024.103372
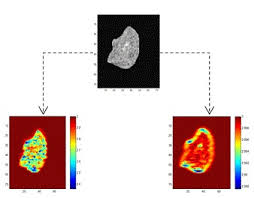
5) Sunday (May 4, 2025) presented by Raneem, “Anatomically plausible segmentations: Explicitly preserving topology through prior deformations”. url: https://doi.org/10.1016/j.media.2024.103222
6) Sunday (May 18, 2025) presented by Alaa, “DeepFake-Adapter: Dual-Level Adapter for DeepFake Detection”. url: https://doi.org/10.1007/s11263-024-02274-6
(GoogleMeet link: https://meet.google.com/wah-nwrt-pqv)
6) Sunday (Jan. 12, 2025) presented by Fatima, “DAEGAN: Generative adversarial network based on dual-domain attention-enhanced encoder-decoder for low-dose PET imaging”. doi: 10.1016/j.bspc.2023.105197.
(GoogleMeet link: https://meet.google.com/wah-nwrt-pqv)
(GoogleMeet link: https://meet.google.com/wah-nwrt-pqv)
(GoogleMeet link: https://meet.google.com/wah-nwrt-pqv)
(GoogleMeet link: https://meet.google.com/wah-nwrt-pqv)
5) Sunday (May 7, 2023) presented by Abdel Rahman, “ViXNet: Vision Transformer with Xception Network for deepfakes based video and image forgery detection. doi.org/10.1016/j.eswa.2022.118423.